Overcoming ambiguity in human-generated data
When the future of data analysis is getting interesting
tl;dr
It’s hard to get structured data from messy text.. we’re gonna see how new functions (AI.GENERATE_TABLE and AI.GENERATE) use AI to easily extract and structure this data directly in BigQuery, simplifying analysis
Messes are made of information
If there’s one thing everyone seems to agree on, one thing that seems common to all countries around the globe, it is how difficult medical indications are to read. Leaving aside the potentially dubious legibility, it’s primarily a cryptic language for the Muggles that we are. It’s a reminder that, more generally, declarative data is always unstructured and it can be hard to extract valuable information from it. The first step is therefore always to find a way to succeed in extracting structured information: a clever regex, a manual decision tree are on the table but overall, simple options are rare. At least it was the case before .. the GenAI wave (or tsunami).
Extracting structured information never has been so easy - if we managed to control the non-deterministic inherent nature of LLMs.
For instance, let’s imagine you are a data-oriented person working in an under-staffed hospital / clinic, you somehow get your hand on various transcripts. Lucky you, it’s already digitized.
Before anything, let’s create a GCS bucket and upload our CSV file on it using the Cloud Shell
gcloud storage buckets create --location=europe-west9 gs://mfg-transcripts
gcloud storage cp medical-transcripts.csv gs://mfg-transcripts
Next, we’ll set up the necessary structure in BigQuery and load the data. We can achieve this all in SQL, executed via the BigQuery UI’s query editor:
CREATE SCHEMA PrincetonPlainsboro
OPTIONS (
description = 'Anonymised patient data',
location = 'EUROPE-WEST9'
);
LOAD DATA OVERWRITE PrincetonPlainsboro.MedicalTranscript
(transcript STRING)
FROM FILES (
format = 'CSV',
uris = ['gs://mfg-transcripts/medical-transcripts.csv']
);
Ok, it’s quite straightforward. Now, let’s examine the data.
As shown before, it is (almost) structured and we can quickly find an extraction pattern. Until recently I would have come up with some regex to extract relevant information. Something like this :
WITH MedicalTranscript AS (
SELECT 'Patient ID: 3780-Lambda4. Age: 12. Marital Status: Single. Diagnosis: Persistent Allergic Rhinitis. Prescription: Fluticasone propionate nasal spray, one spray per nostril daily; Cetirizine 10mg oral tablet, once daily.' AS transcript
)
SELECT
REGEXP_EXTRACT(transcript, 'Patient ID: ([^.]+)') AS PatientID,
REGEXP_EXTRACT(transcript, 'Age: ([^.]+)') AS Age,
REGEXP_EXTRACT(transcript, 'Marital Status: ([^.]+)') AS MaritalStatus,
REGEXP_EXTRACT(transcript, 'Diagnosis: ([^.]+)') AS Diagnosis,
REGEXP_EXTRACT(transcript, 'Prescription: ([^.]+)') AS Prescription
FROM MedicalTranscript;
However, real-world datasets often contain inconsistencies. Some records have different structures and lack the precise ‘PatientID:’ substring that we aim to leverage with REGEXP_EXTRACT. If these problematic records constituted less than 5% of the dataset, manual cleanup (or excluding them and calling it a day) could be a viable workaround. But if this percentage were higher .. we would have a whole other kind of problem on our hands.
Fortunately, this type of problem is one area where generative AI really shines : a simple prompt “From the provided transcript, extract the following details if available: PatientID, Age, Marital Status, Diagnosis, and Prescription.” will get us the structured data we crave. We could run the prompt using the BQML GENERATE_TEXT function, ask for a JSON-structured output, and finally extract each piece of information. This would work perfectly, but there is now a simpler way to navigate that : let me introduce to you the promising AI.GENERATE_TABLE
The goal of this great feature is to easily leverage a LLM and then format the model’s response using a SQL schema, while doing it at BigQuery scale. The only prerequisite is to create a remote model in our dataset, you can use a custom configuration_id but I’ll go with the default one for the sake of simplicity.
CREATE OR REPLACE MODEL `PrincetonPlainsboro.gemini-flash-model`
REMOTE WITH CONNECTION DEFAULT
OPTIONS(ENDPOINT = 'gemini-2.0-flash-001');
And that’s pretty much the only thing you need to configure. From now on, you can very simply extract structured data from your medical transcript by specifying the expected data schema and create a new table based on that.
CREATE TABLE PrincetonPlainsboro.StructuredMedicalTranscript AS
SELECT
PatientID,
Age,
MaritalStatus,
Diagnosis,
Prescription
FROM
AI.GENERATE_TABLE(
MODEL `PrincetonPlainsboro.gemini-flash-model`,
(SELECT transcript AS prompt FROM PrincetonPlainsboro.MedicalTranscript),
STRUCT("PatientID STRING, Age INT64, MaritalStatus STRING, Diagnosis STRING, Prescription STRING " AS output_schema,
0.1 AS temperature,
0.5 as top_p)
);
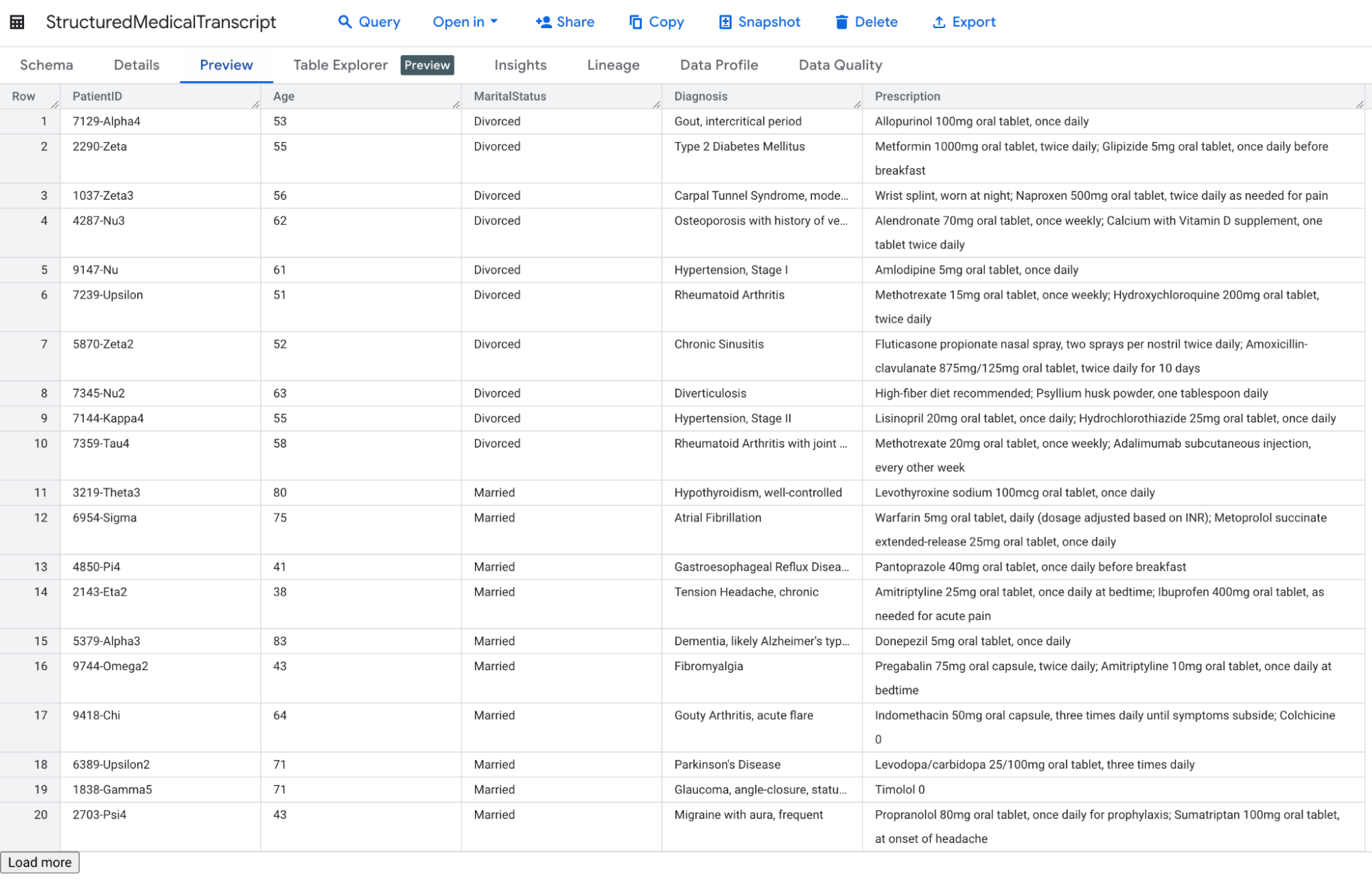
We can even go one step further by enriching our newly created table and try to classify each diagnosis using the AI.GENERATE function, which lets you analyze text in a table. It will later on greatly facilitate visualization. This time, you’ll need to create a connection, but once it’s done, you can write your best prompt at the row level (and because prompt design can strongly affect the responses returned by the model, I strongly advise you to read the Introduction to prompting!).
In our example, that would make a lot of sense to attempt classifying each diagnosis by Body System or Specialty using categories like: Cardiovascular, Endocrine, Musculoskeletal,
To ensure the classification is somewhat deterministic, I will constrain the choices directly within the prompt: “Assign the diagnosis ‘[Insert Diagnosis Here]’ to one of the following categories: Cardiovascular, Endocrine, Musculoskeletal, Neurological, Gastrointestinal, Respiratory, Ophthalmologic, or Other. Output only the category name as plain text.”
SELECT
category,
COUNT(*) AS count
FROM (
SELECT
*,
AI.GENERATE(
('Assign the diagnosis "', diagnosis, '" to one of the following categories: Cardiovascular, Endocrine, Musculoskeletal, Neurological, Gastrointestinal, Respiratory, Ophthalmologic, or Other. Output only the category name as plain text.'),
connection_id => 'eu.gemini-connection',
endpoint => 'gemini-2.0-flash').result as category
FROM PrincetonPlainsboro.StructuredMedicalTranscript
) GROUP BY 1 ORDER BY 2 DESC
And just like that, we have now established a dimension suitable for visualizing the previously unstructured data.
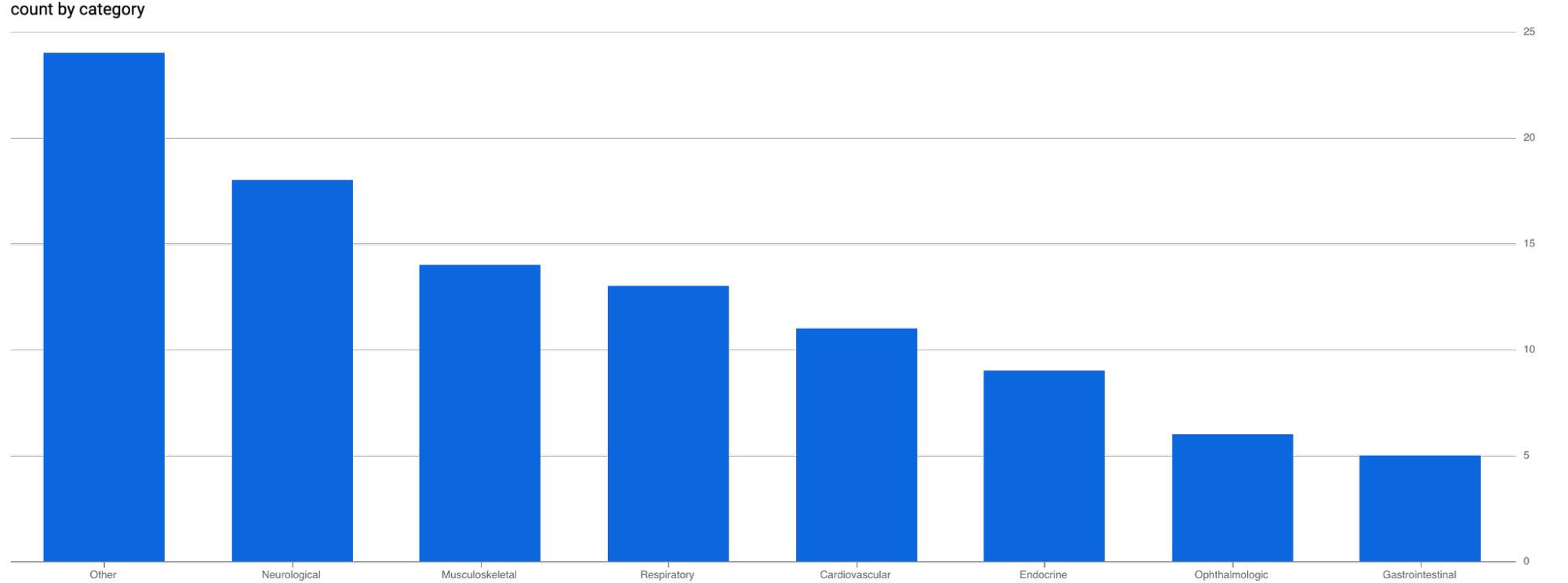
Given that roughly 25% of diagnoses landed in the ‘Other’ category, it surely means I should spend some time improving the category definitions. However, even this initial result from our brief walkthrough demonstrates the considerable potential of BigQuery’s latest AI capabilities. Unstructured data has traditionally been challenging to parse consistently, making the extraction of valuable information a persistent hurdle in data analysis. Although the potential insights were present, accessing them was often impractical.
BigQuery’s native AI functions, specifically AI.GENERATE_TABLE and AI.GENERATE, now offer a powerful and streamlined solution to decipher unstructured text directly within the data warehouse environment. As demonstrated, with minimal setup, free-form text can be converted into structured tables, simplifying previously complex tasks and unlocking new analytical possibilities.
And we can come up with a lot of situations where the core problem is the lack of predefined format and the inherent variability and ambiguity of human-generated content : Customer Service Call Transcripts/Recordings, Social Media Feeds for Brand Monitoring, Legal Document Review, Handwritten Maintenance Logs or Field Reports from Industrial companies, Analyzing Open-Ended Survey Responses.
This opens and facilitates a lot of potential new use cases - the future is looking interesting!
One last thing before you go: given the non-deterministic nature of LLMs, the aforementioned functions are better suited for analytical purposes rather than deployment in production workloads (and AI functions should never justify replacing a deterministic approach merely because the implementation is too complex). More generally, a new wave of native AI functions is coming in the upcoming months but we’ll cover that in more detail when the time comes.